Thresholding Confidence
Introduction
When thresholding outlier likelihood scores, we obtain a binary array of thresholded labels that categorize data points as either inliers or outliers. However, to confidently assess whether these thresholded labels are within an acceptable range of thresholding confidence for their assignments, a statistical approach is required.
There are various methods to approach this, and one such method involves the use of t-test confidence intervals. A t-test is a statistical test used to compare the means of a sample or two groups of samples. It serves as a hypothesis test to determine whether a point within a sample or two groups of samples differs significantly from one another and quantifies this difference. A confidence interval refers to the probability that a population parameter will fall within a set of values for a certain proportion of times. These confidence intervals can be employed to evaluate whether a data point falls within or outside a specific level of confidence in a sample’s distribution. This clear boundary can help determine if a data point is statistically significant in its assigned label.
In the context of outlier detection thresholding, this ‘sample’ refers to the assigned label and can be used to quantify, with a selected degree of confidence, whether the assigned label is statistically significant. If a data point falls outside this confidence interval, it suggests that the data point is more accurately attributed to a new label, signifying uncertainty regarding its status as an inlier or outlier
In order to determine this thresholding confidence for thresholding
outlier detection likelihood scores as mentioned above, the CONF
utility in pythresh.utils.conf has been specifically written for
this purpose.
Methodology
Before jumping into how the thresholding confidence is calculated, it is
important to note that PyThresh essentially has two fundamental
types of thresholding methods. First, are methods that have precise
points within the outlier likelihood scores where a threshold point is
set. These methods can be referred to as continuous based methods. The
other thresholding methods rather define inliers and outlier based on
some classification type criterion and therefore possess no defined
threshold point. These methods can be referred to as classification
based methods.
Continuous Based
For continuous based thresholding methods, the thresholding confidence is calculated as follows:
All the outlier likelihood scores are thresholded and the
.thresh_attribute is calculatedA sample of outlier likelihood scores is selected.
The chosen thresholding method is then applied to this sample.
The
.thresh_attribute of an evaluated thresholder is then stored, defining the boundary from the sample that defines inliers from outliers.The above three processes are repeated based on the number of chosen tests and each boundary value is stored.
This stored list of boundaries now contains a distribution of boundary points for the selected thresholder.
The upper and lower confidence intervals of this distribution can be calculated using the confidence interval equation for a sample given by
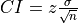 where
where 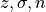 the t-distribution critical value for a selected confidence level
for a sample size, the standard deviation of the distribution, and
the number of datapoints within the sample respectively.
the t-distribution critical value for a selected confidence level
for a sample size, the standard deviation of the distribution, and
the number of datapoints within the sample respectively.With the lower and upper confidence intervals, outlier likelihood scores that fall within the threshold bound with regards to all the outlier likelihood scores
.thresh_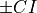 are then set as
uncertains and their indeces are returned.
are then set as
uncertains and their indeces are returned.
Classification Based
For classification based thresholding methods, the thresholding confidence is calculated as follows:
All the outlier likelihood scores are thresholded and their binary labels are stored
A sample of outlier likelihood scores is selected from a stratified list of the above labels.
The chosen thresholding method is then applied to this sample.
The new labels for this sample is stored
The above three processes are repeated based on the number of chosen tests sample labels are stored.
Using the stored 2D array of labels, the ratio for each datapoint based on the number of times it was classed the same as the binary labels for the whole dataset versus the total number of tests is calculated.
From this a two independent sample confidence interval test can be calculated using
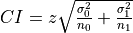 where
where 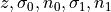 are the t-distribution critical value for a selected confidence
level for the combined sample size, the standard deviation of the
sample for the inlier label ratios, the sample size of the inliers,
the standard deviation of the sample for the outlier ratios, and the
sample size of the outliers respectively.
are the t-distribution critical value for a selected confidence
level for the combined sample size, the standard deviation of the
sample for the inlier label ratios, the sample size of the inliers,
the standard deviation of the sample for the outlier ratios, and the
sample size of the outliers respectively.With the lower and upper confidence intervals, inlier labels ratios that lie beyond the mean of the inlier ratio plus
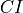 , and
outlier labels ratios that lie beyond the mean of the outlier ratio
minus
, and
outlier labels ratios that lie beyond the mean of the outlier ratio
minus 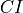 are then set as uncertains and their indeces are
returned.
are then set as uncertains and their indeces are
returned.
Example
Below is a simple example of how to apply the CONF method for the
musk dataset:
import os
import numpy as np
from pyod.models.iforest import IForest
from pyod.utils.utility import standardizer
from pythresh.thresholds.clf import CLF
from pythresh.thresholds.iqr import IQR
from pythresh.utils.conf import CONF
from scipy.io import loadmat
from sklearn.decomposition import PCA
mat_file = 'musk.mat'
mat = loadmat(os.path.join('data', mat_file))
X = mat['X']
y = mat['y'].ravel()
X = standardizer(X)
clf = IForest(random_state=1234)
clf.fit(X)
scores = clf.decision_scores_
thres = IQR()
labels = thres.eval(scores)
confidence = CONF(thres, alpha=0.05, split=0.2)
unc_idx = confidence.eval(scores)
decomp = PCA(n_components=2, random_state=1234)
X = decomp.fit_transform(X)
uncertain = X[unc_idx]
ouliers = X[labels==1]
inliers = X[labels==0]
fig = plt.figure(figsize=(18, 12))
plt.plot(inliers[:, 0], inliers[:, 1], 'y.', label='Inliers', markersize=10)
plt.plot(outliers[:, 0], outliers[:, 1], 'r.', label='Outliers', markersize=11)
plt.plot(uncertains[:, 0], uncertains[:, 1], 'b.', label='Uncertains', markersize=12)
plt.legend()
plt.show()
Below are two scatter plots of the results from the example code above.
However, in the second plot the use of a classification type thresholder
CLF has been employed.
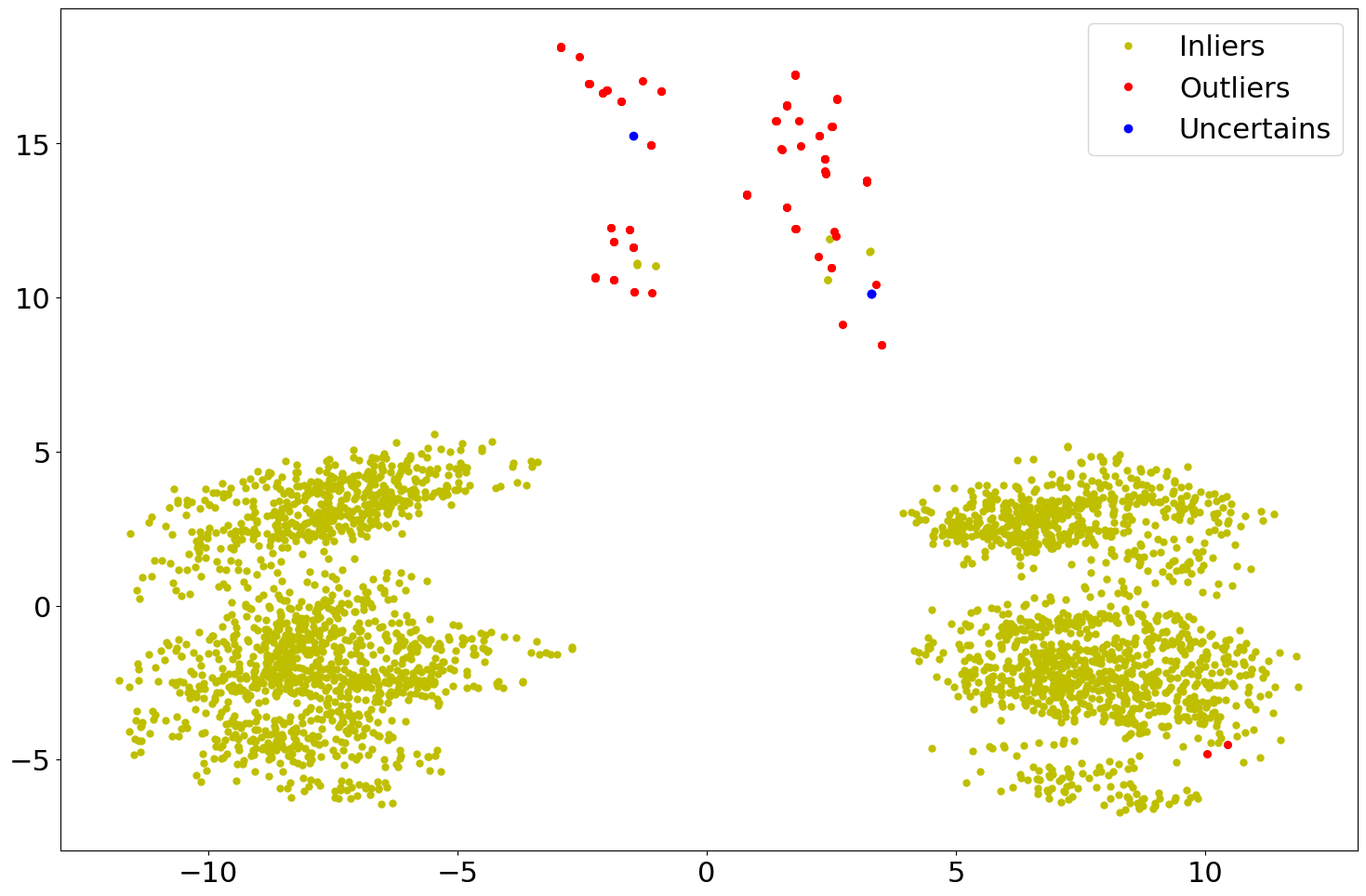
Figure 1: Scatter plot of the CONF evaluated results using IQR.
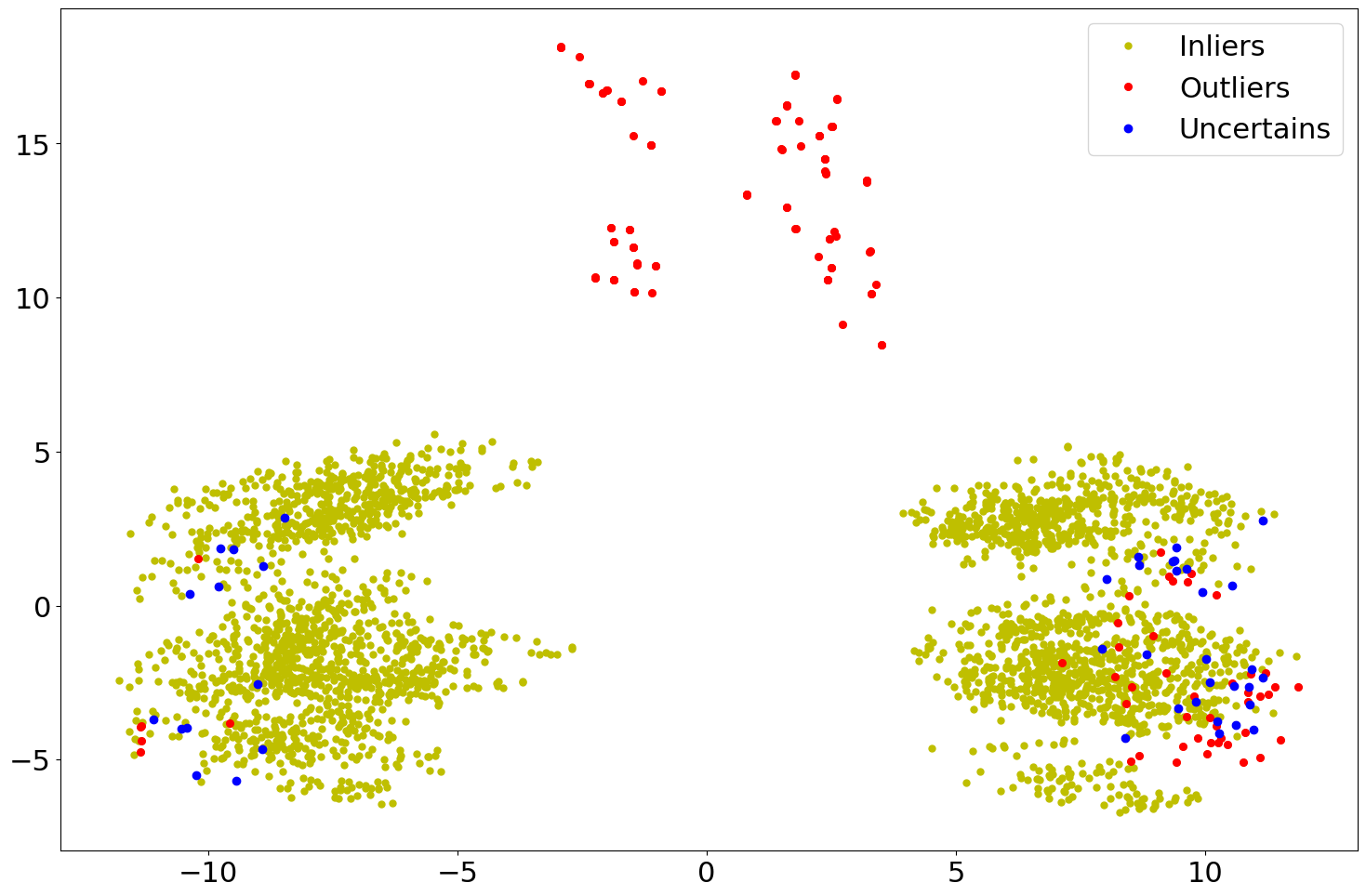
Figure 2: Scatter plot of the CONF evaluated results using CLF.